de novo Pyrimidine Biosynthesis: Bacillus subtilis (WIT2)
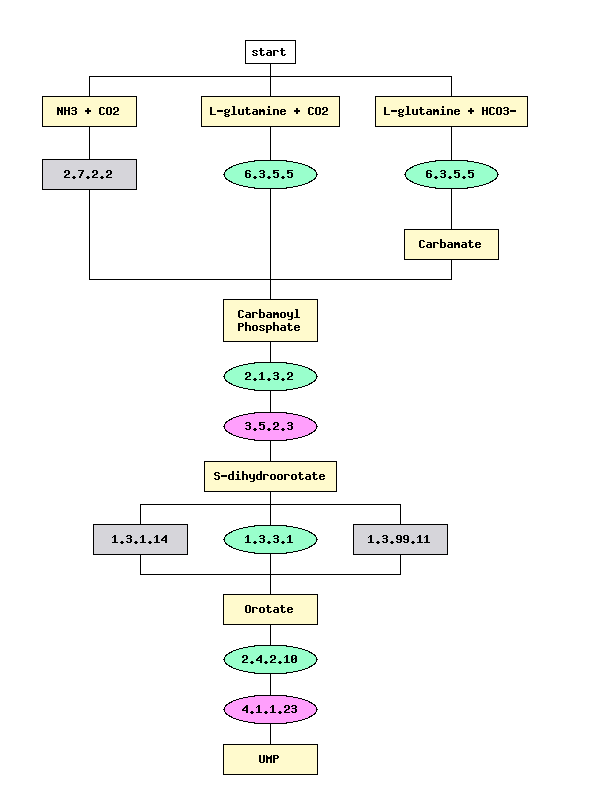
Figure 3.
Figure 3 represents a pathway of 'de novo' Pyrimidine biosynthesis. Alternative
enzymes performing some of the steps in this pathway used by different
organisms are also presented. (For ex: three alternative enzymes are catalyzing
conversion of S-dihydroorotate to orotate, namely, EC 1.3.1.14 - orotate
reductase (NADH), EC 1.3.3.1 - dihydroorotate oxidase and EC 1.3.99.11
- dihydroorotate dehydrogenase). Color coding represents the results of
automated metabolic reconstruction of this pathway for B.subtilis.
Green color signifies the enzymes for which the corresponding genes were
found in the genome; grey color the enzymes that were not found. Pink color
marks the signature enzymes that are unique to a given process and were
identified in B. subtilis. Oval shape marks the enzymes for which
corresponding genes are forming conserved chromosomal gene clusters.
Color and shape scheme:
-
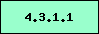 Present in this
organism, NOT a signature enzyme
Present in this
organism, NOT a signature enzyme
-
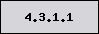 Absent in this organism,
NOT a signature enzyme
Absent in this organism,
NOT a signature enzyme
-
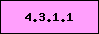 Present in this organism,
signature enzyme
Present in this organism,
signature enzyme
-
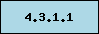 Absent in this
organism, signature enzyme
Absent in this
organism, signature enzyme
-
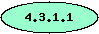 The oval shape marks
enzymes in which at least one of the subunits is a part of a conserved
chromosomal gene cluster.
The oval shape marks
enzymes in which at least one of the subunits is a part of a conserved
chromosomal gene cluster.


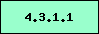 Present in this
organism, NOT a signature enzyme
Present in this
organism, NOT a signature enzyme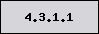 Absent in this organism,
NOT a signature enzyme
Absent in this organism,
NOT a signature enzyme Present in this organism,
signature enzyme
Present in this organism,
signature enzyme Absent in this
organism, signature enzyme
Absent in this
organism, signature enzyme The oval shape marks
enzymes in which at least one of the subunits is a part of a conserved
chromosomal gene cluster.
The oval shape marks
enzymes in which at least one of the subunits is a part of a conserved
chromosomal gene cluster.